| PAPER |
|---|
| mixSeq: A Simple Data Augmentation Method for Neural Machine Translation |
| SeqMix: Augmenting Active Sequence Labeling via Sequence Mixup |
MixSeq in Machine Translation
Most data augmentation techniques operate on a single input, which limits the diversity of the training corpus. MixSeq in machine translation is a simple but effective augmentation method: randomly sample two aligned sequence pairs, and then concatenate their source sentences and the target sentences respectively with a special label <sep> separating two samples.
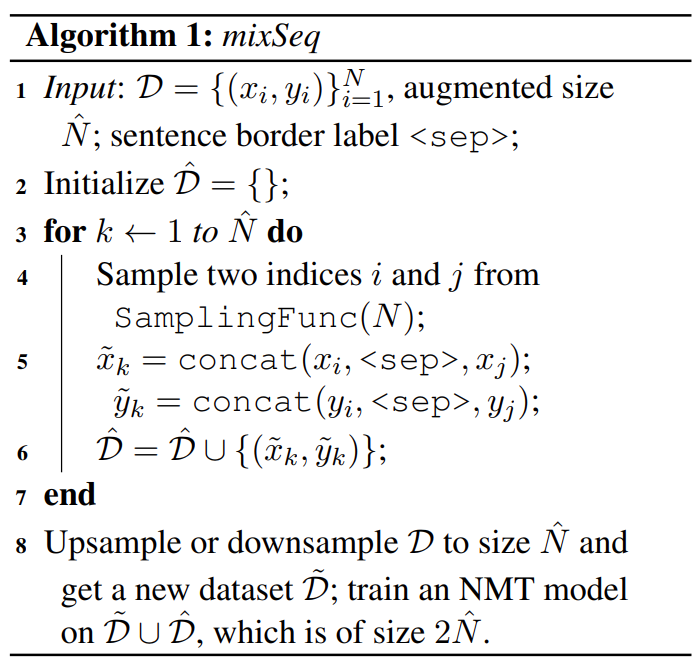
Notice <sep> helps the model learn to align each part of the input to the corresponding part of the output, which can improve the representation learning. Concatenating in translation helps $x_i$ pay more attention to $y_i$, instead of $y_j$. It is like contrastive learning which introduces negative samples to improve performance. If the two concatenated sequences are contextually related, we can enhance MixSeq to a context-aware version: ctxMixSeq, which only samples consecutive sequences in a given document. MixSeq can also be used with other conventional data augmentation methods.
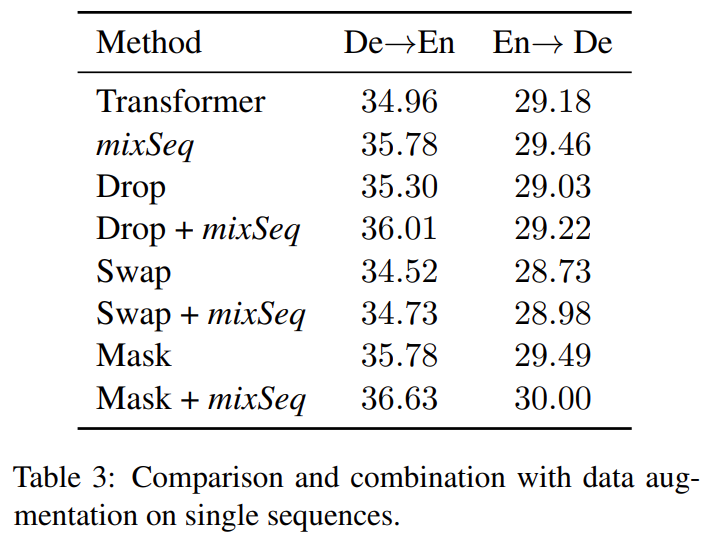
Alignment in sequence mixup is important, where we need to design a proper sampling function; concatenating more sequences should be also considered.
MixSeq in Active Sequence Labeling
Active sequence labeling starts with a small labelled corpus and a query policy. A fixed number of unlabelled sequences are selected by the query policy for annotation and then model updating. Current active sequence labeling methods just use the queried samples alone, to which MixSeq can also be applied.
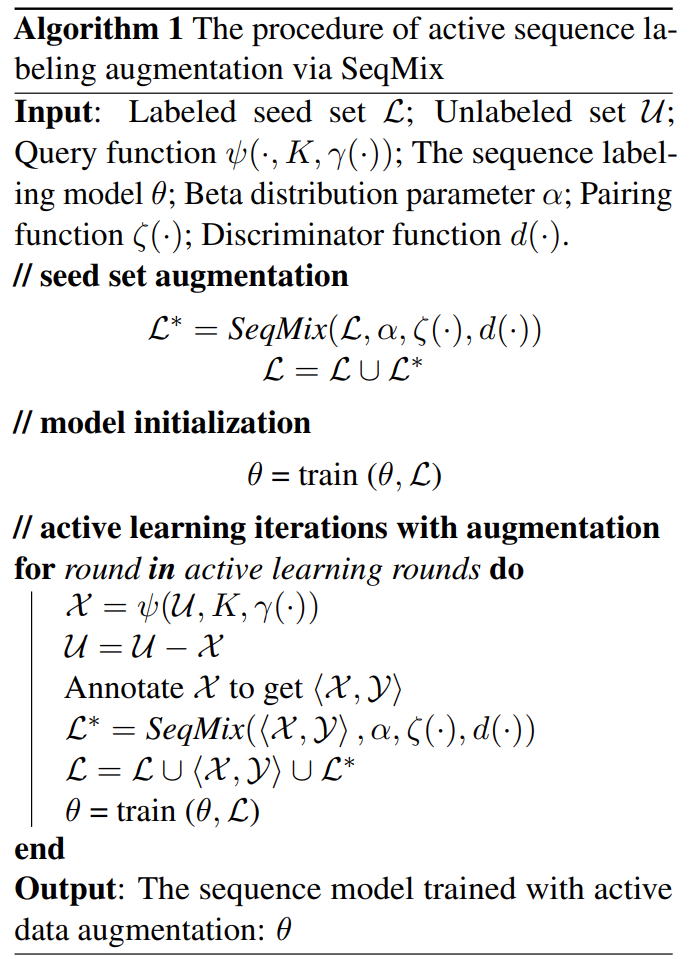
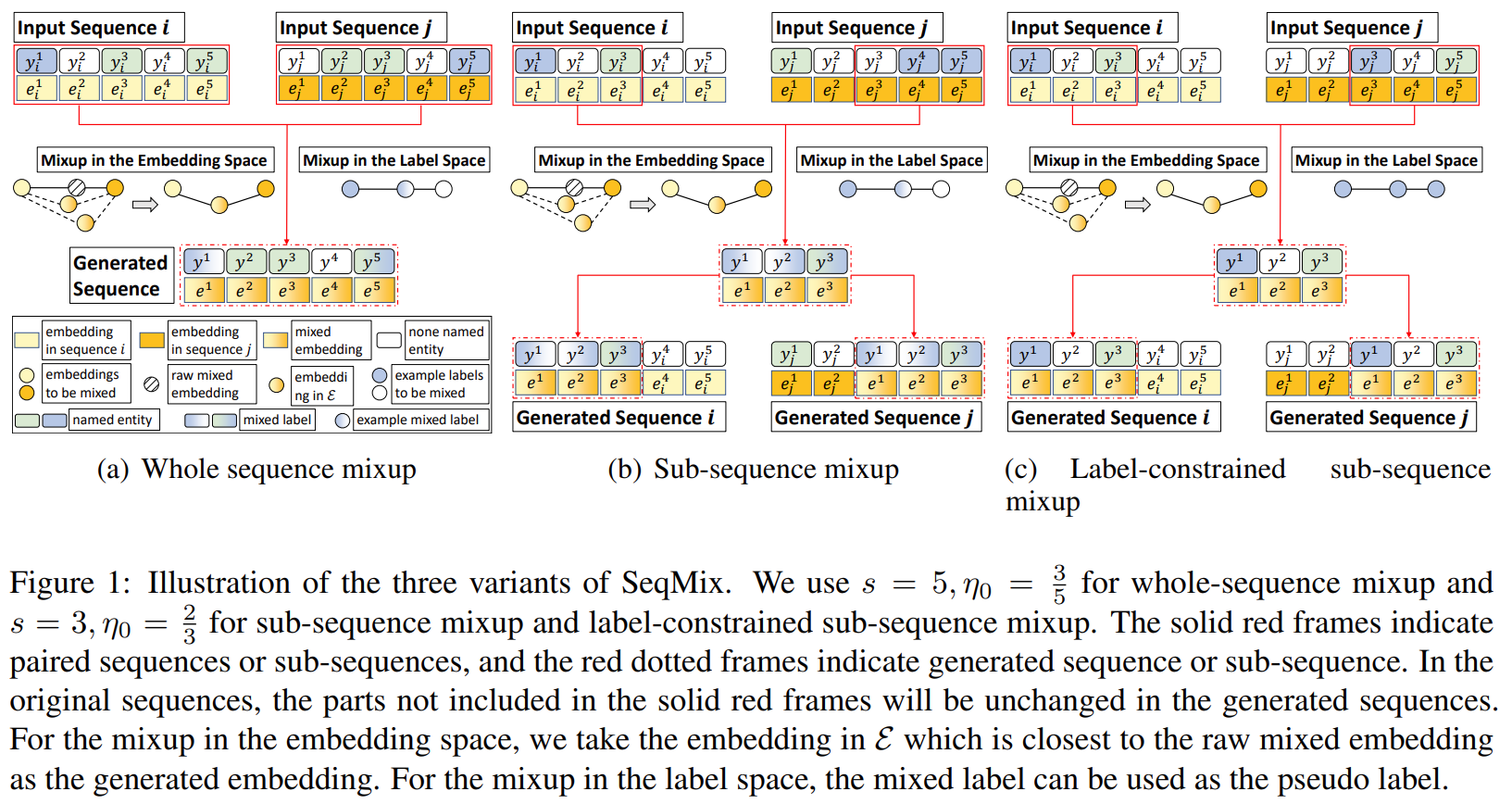
MixSeq in active sequence labeling performs mixup for both sequences and token-level labels of the queried samples. Unlike previous MixSeq, there is a discriminator judges whether the generated sequences are plausible or not. The discriminator will select the low-perplexity sequences as plausible ones.
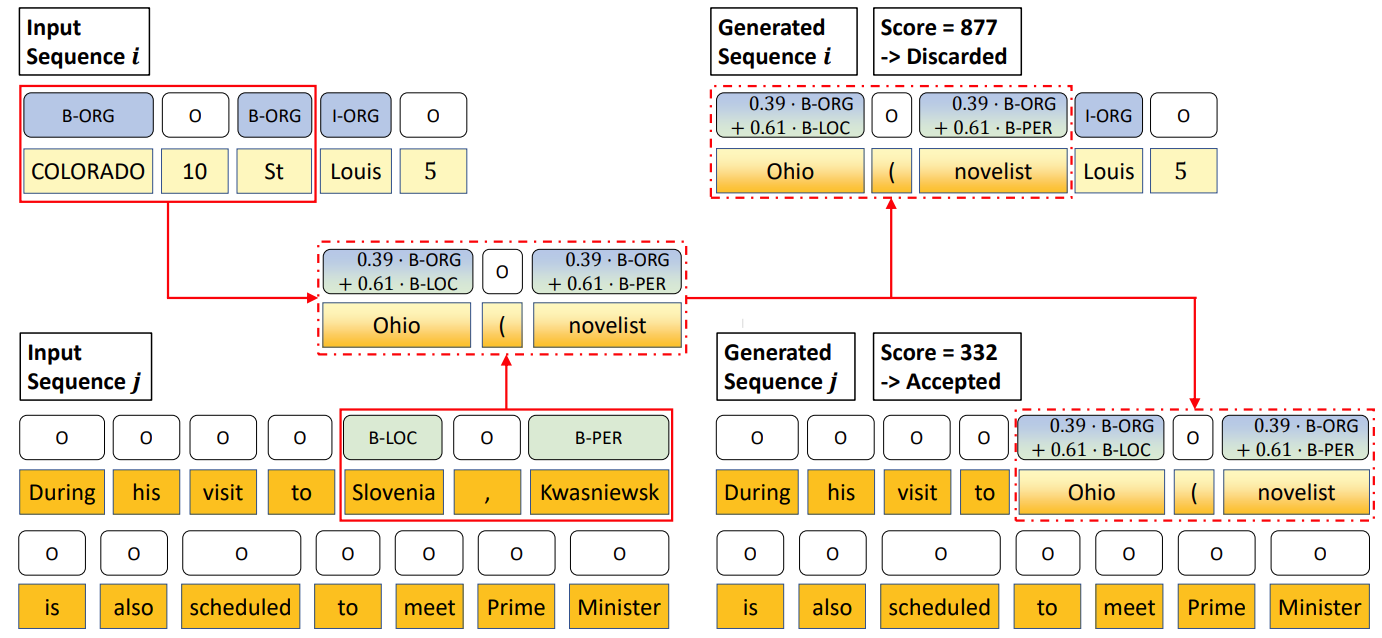
Ablation study on MixSeq in active sequence labeling is shown as follows. Notice in many sequence labeling tasks, the labels of interest are scarce. Thus, we should design a sequence pairing function to select more informative parent sequences for mixup (mixup dominating factors). The best MixSeq method is sub-sequence mixup with NTE (Normalized Token Entropy) sampling.